Research - (2022) Volume 12, Issue 10
Phenotypic and genotypic characterization of lactococci isolated from different kinds of raw milk (goat, cow, sheep and camel)
K. Houbad1*, A.M.A. Bekada1, A. Homrani1 and Y. Djellid2,3Abstract
Lactococci were isolated from different types of raw milk (goat, cow, sheep and camel) from different regions of Algeria.
From a collection of 254 cocci isolates, only 110 strains representative of the dominant flora were completely identified, by phenotypic methods.
Phenotypic and genotypic identification of these isolates was performed using classical biochemical and physiological methods then by sequencing of the 16S rDNA gene.
A molecular analysis was conducted where the phylogenetic tree was constructed using the Neighbor-Joining method.
The results revealed a dominance of the species Lactococcus lactis subsp. Lactis with a percentage of 41%; followed by Lactococcus lactis subsp. lactis. biovar. diacetylactis (12%), then Lactococcus lactis subsp. hordniae (11%), Lactococcus lactis subsp. Cremoris, Lactococcus plantarum and Lactococcus raffinolactis (10%), and lastly,Lactococcus lactis Subsp.tructae.,lactococcus hircilactis, lactococcus lendensis and lactococcus piscium are found with very low percentage.
Keywords
Milk, Lactococci, Identification, Molecular tree, 16S rDNA gene, Neighbor-joing method.
Introduction
The food industry uses a number of processing aids, including lactic acid bacteria.
They are involved in the dairy industry and in the fermentation of many other food products, contributing to the texture and flavor of foods and to the production of aromatic compounds. They ferment carbohydrates into lactic acid, which lowers the pH for the bio-preservation of foodstuffs.
The interest of lactic acid bacteria in fermented foods lies mainly in certain technological properties, namely, the acidifying power that has an important influence on the formation of coagulant and gelation, as well as the flavoring power, the texturing power, the proteolytic and lipolytic activity that have a well determined role on the organoleptic quality of the fermented food (Randazzo et al., 2009).
Since ancient times, lactic acid bacteria have been used for food manufacturing and preservation (Chammas et al., 2006; Zamfir et al., 2006), as they are generally recognized as safe, of "GRAS" (Generally Recognized As Safe) status (O'Sullivan et al., 2002).
Today, lactic acid fermentation is a growing activity in both industrial and artisanal sectors. Several microorganisms are involved in the manufacture of fermented dairy products (Johnson and Steele, 2001).
In the dairy industry, lactic strains are selected based on their technological properties (lactic acid production, aroma production, proteolytic activity and growth kinetics), and their functional characteristics (antibacterial activity) (Tamime, 2002; Molin, 2008); satisfying then the sanitary needs in food industry and to inhibit the proliferation of pathogenic microorganisms (Ross et al., 2002).
Materials and Methods
Sampling
A total of 20 samples of raw milk of camel, ewe, goat and cow was collected fromhealthy animals in different regions in Algeria (Table 1).
| Samples | Number of samples | Collection period | Region |
|---|---|---|---|
| Cow’s milk | 4 | 2016, 2017, 2018 | Tiaret, Sidi Bel Abbes, Relizane |
| Sheep’s milk | 4 | ||
| Goat’s milk | 5 | ||
| Camel milk | 7 | Ghardaïa, Béchar, El-bayadh, Djelfa, M’sila |
Table 1. Milk samples.
The milk samples were collected in sterile 250 ml glass vials, transported in refrigerated coolers, and stored at 4°C then analyzed within 24 hours.
Preliminary Analyses
pH test
The measurement of pH was carried out by pH meter (Inolab MLM). The reference electrode for the measurement of the concentration of H+ions (thus the pH) is the hydrogen electrode. This specially treated platinum electrode is immersed in the solution whose pH is to be measured (Lehninger, 1981).
Reductase test
The reductase test is used to estimate the microbial load of milk samples. Its principle is based on the decoloration of methylene blue. The speed of this discoloration is directly proportional to the number of germs present (Larpent et al., 1997). The higher the microbial activity, the shorter the duration of decoloration of samples (Ryffel, 2007).
Isolation and purification of lactic acid bacteria
Isolation
Most bacteria that grow in milk can be isolated using conventional microbiological techniques, such as culture on suitable nutrient agar medium (Federico, 2012).
The selective isolation of lactococci is performed by culture on Elliker selective medium (Elliker et al., 1956): tryptone 20 g, yeast extract 5 g, gelatin 2.5 g, NaCl 4 g, sodium acetate 1.5 g, glucose 5 g, sucrose 5 g, lactose 5 g, ascorbic acid 0.5 g, agar-agar 15 g, 1000ml distilled water, pH=7, autoclaving 120°C for 20minute.
Depending on their concentration, a serial dilution of the sample in sterile physiological water (0.9% NaCl) is necessary before cultivation on Elliker medium; of which we inoculated 1 ml of each dilution by spreading in depth. The plates were incubated at 30°C for 24 to 48 hours.
Purification
After growth, whitish or milky colonies were subjected to preliminary tests including Gram stain and catalase test (Badis et al., 2006). Only Gram-positive and catalase-negative strains were retained and transferred to liquid Elliker medium and incubated at 30°C for 24 hours. After incubation, the appearance of cloudiness indicates thebacterial growth; 0.1 ml of each positive tube is streaked on solid Elliker. Incubation at 30°C for 24 hours. The operation is repeated until a pure culture is obtained. The purity of the strains is revealed by homogeneous colonies having the same external aspect (color, size and shape) (Guiraud, 2004).
Conservation of the strains
The strains were cultured on Elliker agar slants in tubes, then conserved at 4°C and the transplants were performed every two weeks. For long term conservation, a culture of 18 hours was centrifuged (4000 rpm during 10 min). Then the pellet was washed and transferred to skimmed milk added with 30% glycerol and 1% yeast extract. The culture was stored at-20°C (Samelis et al., 1994 and Herrero et al., 1996).
Identification of lactococci
Phenotypic characterization
The bacterial isolates were characterized using phenotypic methods to verify that the isolates are lactic acid bacteria.
Gram-positive, catalase-negative isolates were analyzed at the genus level.
The identification was established based on morphological characters (macroscopic study: (size, color, shape, appearance), microscopic study: (Gram, cell morphology, mode of association) and various physiological and biochemical characters:
• Sherman test and thermoresistance at 60.5°C for 30 min (Samelis et al. 1994).
• Fermentation type: This test allows defining the type of metabolism by which the carbon substrate is transformed. It consists in highlighting the fermentation of gas (CO2). Thus lactic bacteria can be classified as homo or hetero-fermentative on liquid Elliker with a Durham bell to notice the production of CO2. After incubation at 30°C for 48 hours, the presence or absence of gas in the bell indicates the fermentative type (Kihal, 1996; Hariri et al., 2009).
• Growth on Elliker medium was monitored for temperatures: 10°C after incubation for 5,7 days) and 40°C, 45°C after incubation for 24h to 48h (Badis, 2004).
• Production of arginine dehydrolase on the M16BCP medium of Tomas (1973).
• Growth in Elliker alkaline pH=9.6 and acidic pH=4.
• Growth in the presence of different concentration of sodium chloride NaCl on hypersaline bolin with 4% and 6.5% NaCl.
• Production of acetoin on the Clark and Lubs medium (Samelis et al., 1994; Guiraud, 1998).
• Hydrolysis of esculin on BEA medium (Dellaras, 2007).
• Use of citrate in the presence of fermentable sugar (glucose):
• The use of citrate is studied on Kempler and MC KAY (1980) medium.
• Use of carbohydrates: The fermentation of carbohydrates was conducted on medium without meat extract and added bromocrysol purple (BCP) as a pH indicator, whose only carbon source would be one of the sugars: arabinose fructose, lactose, maltose, mannitol, raffinose, rhamnose, sucrose and D-xylose.
Genotypic trait
During the last decades, different methods of molecular identification of lactic acid bacteria have been developed in order to reduce the analysis time.
DNA extraction
Most protocols for the preparation of bacterial genomic DNA consist of lysis, followed by incubation with a nonspecific protease and a series of extractions before precipitation of the nucleic acids. Such procedures effectively remove contaminating proteins, but are not effective in removing exopoly-saccharides that may interfere with the activity of enzymes such as restriction endo-nucleases and ligases.
DNA extraction and purification was performed by the CTAB/NaCl method described by Wilson (1987), and modified using 1mg/mL lysozyme (BIOMATIK) for cell wall digestion (Fhoula et al; 2013).
After a fresh culture of the strains to be identified for 18 hours on Elliker medium at 30°C, we go through the following steps:
1. Centrifugation (12000 rpm) for 15 min and recovery of the bacterial pellet of 1.5 ml or 3 ml.
2. Wash the pellet with 0.5 ml of sterile water.
3. Re-suspend in 550 μl of sterile water.
4. Add 17 μl lysozyme (35 mg/ml) and incubate at 37°C for 30min.
5. Add 6 μl Proteinase K (20 mg/ml)+30 μl SDS (10%) and incubate at 37°C for 30 min.
6. Add 100 μl NaCl (SM)+80 μl CTAB and incubate at 65°C for 10min.
7. Adding 700 μl CAI (chloroform 24 ml/iso-amyl alcohol 1 ml) inversion then centrifugation (10000 rpm/10 min).
8. Collect the supernatant using a new ependorff.
9. Repeat the 7ème and 8ème steps.
10. Add 800 μl of 97° alcohol and incubate for 2 hours at (-20°C).
11. Cold centrifugation (8000 rpm/30 min).
12. Pouring the supernatant gently.
13. Drying for 30 min at 37°C.
14. Re-dissolve the pellet in 30 μl of sterile water.
15. Storage at -20°C.
DNA amplification (PCR: Polymerase Chain Reaction)
PCR amplification assays were performed with a Biorad Icycler thermocycler (Biorad, USA) with the universal primers described by LANE (1991) synthesized by Eurofins Genomics, (France)
• 27f (forward): 5' AGAGTTTGATCCTGGCTCAG3'.
• 1492R (Reverse): 5' GGTTACCTTGTTACGACTT 3'.
These primers allow to amplify the 16SrDNA gene (1.5 Kb) approximately.
DNA amplification reactions were performed in a 25 μl volume containing 1X reaction buffer, 1.5 mM MgCl2, 0.2 mM dNTPs mixture, 0.5 ��M of each primer, 1UT aqpolymerase (Fermentas), and 150 ng of total DNA, using the following program: 94°C for 5min(initial denaturation), followed by 35 cycles of 94°C for 45S (denaturation), 55°C for 1 min (hybridization) and 72°C for 2 min (elongation), and a final extension step at 72°C for 7 min(final elongation).
Stoker before development at 10°C until use.
To verify the presence of a good amplification of the target gene, the PCR products are deposited on a 1.5% agarose gel in an electrophoresis tank containing 0.5% TBE (Tris, Borate, EDTA) buffer. The migration is done for 30 min. The DNA was visualized and photographed under UV.
Sequencing
Sequencing of 16S ribosomal DNA) was performed using the universal primers: 27F and 1492R.
Sequencing of the amplified and purified DNA was performed by Eurofins genomic (France) according to the Sanger method (Sanger et al., 1977) using the BigDye® Terminator v3.1 cycle sequencing kit (Applied Biosystems) and universal primers (27F and 1492R).
The sequences obtained are then compared with those of the GeneBank database on the NCBI site (National Center for Biotechnology Information).
The molecular phylogeny of the 16S rDNA gene
We performed phylogenetic analysis using 16S rDNA sequences by MEGA version 6 software (Tamura et al., 2013), according to the Neighbour-Joining method (Saitou and Nei, 1987). For this purpose, we performed sequence alignment. In addition to the DNA sequences resulting from our sequencing, so-called reference sequences were included to allow for better taxonomic characterization. The sequence of the gene coding the 16S rRNA of the Bacillus sublitis strain was added to serve as an outgroup, for the rooting of the tree (Table 2).
| Strain name 16S rRNA gene ID |
|---|
| CP064339.1 Lactococcus lactis subsp. Lactis |
| MF354327.1 Lactococcuslactis.sp |
| KP764102.1 Lactococcus lactis subsp. lactis |
| MT185438.1 Lactococcus lactis subsp. lactis bv. diacetylactis |
| AM406671 Lactococcus lactis subsp. cremoris |
| MT573739 Lactococcus lactis subsp. hordniae |
| X60646.1 Bacillus.subtilis 16S ribosomal RNA |
Table 2. Reference strains used as guide sequences to establish the phylogenetic tree.
Results and Discussion
All 20 samples from different dairy animals (camel, goat, sheep, cow) in different regions of Algeria Table 1 were collected and analyzed to isolate and identify strains belonging to the genus Lactococcus after milking performed according to different hygienic methods.
The pH varies according to the type of milk. For all samples of milk analyzed, the pH values are between (6.3-7).
The decoloration of the methylene blue by the analyzed milk took place after more than 4 hours. This milk is therefore of good bacteriological quality. It contains less than 2.106 germs/ml (Larpent, 1970; Guiraud, 1998). The discoloration of methylene blue is due to bacterial metabolism and its speed is directly proportional to the number of germs.
The results revealed that the methylene blue decoloration time of the samples occurred after 4 hours. This slow reduction of methylene blue is explained by a low microbial load. This is in agreement with the results of Larpent (1997) where the hygienic quality of the collected milk can be considered as good.
Phenotypic identification of strains
After the morphological tests, only Gram positive and catalase negative bacteria that are in the form of isolated shells, pairs or chains are retained.
The 317 lactic isolates from 20 samples of raw camel, goat, sheep and cow milk with a Gram-positive, catalase-negative homofermentative profile from which 254 cocci isolates were selected, from this collection of cocci based on the results of the Sherman blue test, only 110 isolates representative of the dominant flora were completely identified by phenotypic methods.
The phenotypic, biochemical and physiological identification of lactococci shows a significant diversity of species in the genus Lactococcus.
The criteria used to determine the species and subspecies and the results obtained are recorded in Tables 3,4,5,6, followed by the fermentative profiles of the isolated strains in Table 7.
| Origin of the milk | Cow’s milk | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains | Lv11 | Lv6 | Lv50 | Lv9 | Lv63 | Lv3 | Lv81 | Lv43 | Lv32 | Lv24 | Lv60 | Lv91 | Lv17 | Lv54 | Lv98 | Lv78 | Lv53 | Lv6 | Lv62 | Lv15 | Lv44 | Lv76 | Lv29 | Lv16 | ||
| Methylene blue | 1% | + | + | + | + | + | + | + | + | + | + | + | - | - | - | + | + | + | - | - | - | - | - | - | + | |
| 3% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||
| ADH | + | + | + | + | + | + | + | + | + | + | + | - | - | - | + | + | + | + | + | + | + | - | - | +/- | ||
| CIT | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | + | + | + | + | + | + | - | - | - | ||
| ACT | - | - | - | - | - | - | - | - | - | - | - | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| T° | 10°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| 40°C | + | + | +/- | + | + | + | +/- | + | + | + | +/- | + | +/- | + | + | + | +/- | - | - | - | - | + | + | - | ||
| 45°C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||
| pH | 4 | - | - | - | - | - | - | - | - | - | - | - | + | + | + | - | - | - | - | - | - | - | - | - | - | |
| 9.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||
| NaCl | 4% | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - | - | - | - | - | - | + | |
| 6.5% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||
| RES at 63°C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||
| Esculine | - | - | - | - | - | - | - | - | - | - | - | + | + | + | + | + | + | - | - | - | - | - | - | + | ||
| Identified at | Lc.lactis subsp.lactis | lc.platarum | .lactis subsp.lactis biovar.diacetylactis | Lc.lactis subsp.hordniae | Lc.lactis subsp.cremoris | lc.lendensis | ||||||||||||||||||||
Table 3. Biochemical and physiological criteria for species of lactic acid bacteria of the presumed genus Lactococcus isolated from raw cow's milk.
| Origin of the milk | Sheep’s milk | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| strains | Lb22 | Lb4 | Lb65 | Lb89 | Lb16 | Lb45 | Lb19 | Lb25 | Lb66 | Lb4 | Lb1 | Lb73 | Lb80 | Lb92 | Lb46 | Lb9 | Lb21 | Lb65 | Lb14 | Lb49 | Lb30 | |
| methylene Blue | 1% | + | + | + | + | + | + | + | + | + | - | - | + | - | - | - | - | - | - | - | V | V |
| 3% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ADH | + | + | + | + | + | + | + | + | + | - | - | + | - | - | - | + | + | + | + | V | V | |
| CIT | - | - | - | - | - | - | - | - | - | - | - | + | - | - | - | - | - | - | - | - | - | |
| ACT | - | - | - | - | - | - | - | - | - | + | + | + | + | + | + | - | - | - | - | V | V | |
| T° | 10°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | +/- |
| 40°C | + | + | + | + | + | + | + | + | + | + | + | + | + | +/- | - | - | - | - | - | - | - | |
| 45°C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| pH | 4 | - | - | - | - | - | - | - | - | - | - | - | - | + | + | + | - | - | - | - | +/- | + |
| 9.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| NaCl | 4% | + | + | + | + | + | + | + | + | + | - | - | + | + | + | + | - | - | - | - | + | + |
| 6.5% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| RES at 63°C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Esculine | - | - | - | - | - | - | - | - | - | - | - | + | + | + | + | - | - | - | - | - | - | |
| Identified at | Lc.lactis subsp.lactis | Lc.lactis subsp.cremoris | .lactis subsp.lactis biovar.diacetylactis | lc.platarum | Lc.lactis subsp.hordniae | Lc.lactis subsp.tructae | ||||||||||||||||
Table 4. Biochemical and physiological criteria for species of lactic acid bacteria of the presumed genus Lactococcus isolated from rawsheep’s milk.
| Originof the milk | Goat’s milk | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains | Lc6 | Lc55 | Lc89 | Lc46 | Lc79 | Lc3 | Lc32 | Lc60 | Lc85 | Lc26 | Lc22 | Lc1 | Lc72 | Lc16 | Lc15 | Lc29 | Lc43 | Lc65 | Lc33 | Lc13 | Lc9 | Lc7 | Lc12 | Lc59 | Lc74 | Lc81 | Lc25 | Lc93 | Lc95 | Lc47 | Lc19 | Lc6 | |
| methylene Blue | 1% | + | + | + | + | + | + | + | + | + | + | + | + | - | - | - | - | - | - | - | - | - | + | + | + | + | + | + | - | + | + | - | - |
| 3% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ADH | + | + | + | + | + | + | + | + | + | + | + | + | - | - | - | + | + | + | + | + | + | + | + | + | + | + | + | - | V | V | - | - | |
| CIT | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | + | + | + | - | - | + | + | + | + | + | + | + | - | - | - | - | |
| ACT | - | - | - | - | - | - | - | - | - | - | - | - | + | + | + | + | + | + | + | - | - | + | + | + | + | + | + | - | V | V | + | + | |
| T° | 10°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 40°C | + | + | + | +/- | + | + | + | + | + | + | + | +/- | + | + | + | + | + | + | + | - | - | - | - | - | - | - | - | + | - | - | + | +/- | |
| 45°C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| pH | 4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | +/- | - | + | + |
| 9.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| NaCl | 4% | + | + | + | + | + | + | + | + | + | + | + | + | - | - | - | + | + | + | + | - | - | - | - | - | - | - | - | + | - | - | + | + |
| 6.5% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| RES at 63°C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Esculine | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | + | + | + | - | - | - | - | - | - | - | - | + | + | + | + | + | |
| Identified at | Lc.lactis subsp.lactis | Lc.lactis subsp.cremoris | .lactis subsp.lactis biovar.diacetylactis | Lc.lactis subsp.hordniae | Lc.raffinolactis | lc.piscium | lc.hircilactis | lc.platarum | |||||||||||||||||||||||||
Table 5. Biochemical and physiological criteria for species of lactic acid bacteria of the presumed genus Lactococcus isolated from raw goat’s milk.
| Origin of the milk | Camel’s milk | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| strains | Lm95 | Lm90 | Lm5 | Lm36 | Lm85 | Lm76 | Lm54 | Lm17 | Lm19 | Lm23 | Lm29 | Lm26 | Lm49 | Lm39 | Lm9 | Lm7 | Lm82 | Lm64 | Lm66 | Lm57 | Lm60 | Lm33 | Lm38 | Lm25 | Lm20 | Lm12 | Lm11 | Lm96 | Lm1 | Lm44 | Lm42 | Lm72 | Lm55 | ||||
| methylene Blue | 1% | + | + | + | + | + | + | + | + | + | + | + | + | + | - | - | - | - | + | + | + | + | + | - | - | - | + | + | + | + | + | V | - | - | |||
| 3% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| ADH | + | + | + | + | + | + | + | + | + | + | + | + | + | - | - | - | - | + | + | + | + | + | - | - | - | + | + | + | + | + | V | + | + | ||||
| CIT | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | + | + | + | + | - | - | - | + | + | + | + | + | - | - | - | ||||
| ACT | - | - | - | - | - | - | - | - | - | - | - | - | - | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | V | - | - | ||||
| T° | 10°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | +/- | + | + | |||
| 40°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | +/- | + | - | - | - | - | - | - | - | - | ||||
| 45°C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| pH | 4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | + | + | - | - | - | - | - | +/- | - | - | |||
| 9.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| NaCl | 4% | + | + | + | + | + | + | + | + | + | + | + | + | + | - | - | - | - | + | + | + | + | + | + | + | + | - | - | - | - | - | + | - | - | |||
| 6.5% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | - | - | + | - | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| RES at 63°C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| Esculine | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | + | + | + | + | + | + | + | - | - | - | - | - | - | - | - | ||||
| Identified at | Lc.lactis subsp.lactis | Lc.lactis subsp.cremoris | .lactis subsp.lactis biovar.diacetylactis | lc.platarum | Lc.raffinolactis | Lc.lactis subsp.tructae | Lc.lactis subsp.hordniae | ||||||||||||||||||||||||||||||
Table 6. Biochemical and physiological criteria for species of lactic acid bacteria of the presumed genus Lactococcus isolated from raw camel’s milk.
| Sugars | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Mannitol | - | - | + | + | - | + | - | + | + | - |
| Raffinose | + | V | - | + | - | + | + | + | - | - |
| Rhamnose | + | +/- | + | v | v | V | v | +/- | +/- | - |
| Lactose | - | - | + | + | - | + | + | + | + | - |
| Fructose | + | V | V | v | - | + | v | +/- | +/- | - |
| D-xylose | + | + | + | - | - | + | + | - | + | - |
| Arabinose | + | + | + | - | - | + | v | +/- | V | - |
| Sorbitol | + | V | V | v | v | - | v | + | + | - |
| Saccharose | - | + | - | + | + | + | + | + | + | + |
| Maltose | V | + | + | + | - | v | +/- | +/- | + | |
| Galactose | + | + | v | + | - | + | + | + | + | + |
| Mellibiose | - | + | +/- | + | - | + | + | + | - | - |
| Ribose | + | - | +/- | + | - | - | - | - | - | - |
- Lc.lactis subsp.lactis(Lv11, Lv6, Lv50, Lv09, Lv63, Lv3, Lv81, Lv43, Lv32, Lv24, Lv60, Lb22, Lb4, Lb65, Lb89, Lb16, Lb45, Lb19, Lb25, Lb66, Lc6, Lc55, Lc89, Lc46, Lc79, Lc3, Lc32, Lc60, Lc85, Lc26, Lc22, Lc1, Lm95, Lm90, Lm5, Lm36, Lm85, Lm76, Lm54, Lm17, Lm19, Lm23, Lm29, Lm26, Lm49)
- Lc.lactis subsp.cremoris(Lv76, Lv29, Lb4, Lb1, Lc72, Lc16, Lc15, Lm39, Lm9, Lm7, Lm82)
- Lc.lactis subsp.lactis biovar.diacetylactis(Lv98, Lv78, Lv53, Lb73, Lc29, Lc43, Lc65, Lc33, Lm64, Lm66, Lm57, Lm60, Lm33)
- Lc.lactis subsp.tructae(Lb49, Lb30, Lm42)
- Lc.lactis subsp.hordniae(Lm72, Lm55, Lc13, Lc9, Lb9, Lb21, Lb65, Lb14, Lv6, Lv62, Lv15, Lv44)
- Lc.raffinolactis(Lm12, Lm11, Lm96, Lm1, Lm44, Lc7, Lc12, Lc59, Lc74, Lc81, Lc25)
- lc.piscium(Lc93)
- lc.hircilactis (Lc47 andLc95)
- lc.lendensis (Lv16)
- lc.platarum(Lm38, Lm25, Lm20, Lc19, Lc6, Lb80, Lb92, Lb46, Lv54, Lv17, Lv91)
Table 7. Fermentation profile of isolates of the putative genus Lactococcus sp.
The distribution of species in percentages shows the dominance of the species Lactococcus lactis subsp. Lactis with a percentage of 41%; then Lactococcus lactis subsp.lactis. biovar. diacetylactis (12%), followed by Lactococcus lactis subsp. hordniae (11%), Lactococcus lactis subsp. Cremoris, Lactococcus plantarum and Lactococcus raffinolactis (10%), and lastly, the strains Lactococcus lactis Subsp.tructae. Lactococcus hircilactis, Lactococcus lendensis and Lactococcus piscium are found with very low percentage.
According to the analyses, the 110 isolates identified showed an ability to grow at 10°C and 40°C but not at 45°C showing hemolysis y, some isolates grow on 0.1% Sherman's milk and not on 0.3% and also in a 4% hypersaline broth. They do not grow at pH 9.6, they do not survive after heat treatment and they do not grow at 6.5% NaCl except two strains Lactococcus lactis subsp lactis biovar diacetylactis and Lactococcus lactis subsp cremoris isolated from camel milk which are resistant to this salt concentration (Karam, 2006), they have been attached to the genus Lactococcus.
In relation to the DHA test, acetoine production, esculin hydrolysis, citrate utilization and fermentative sugar profile, they were subdivided into species and subspecies: (Badis, 2004; Dicks et al., 1993; Hammes et al., 1992; Holzapfel and Schillinger 1992; Harrigan and Mc Cance, 1976).
The pre-identification of isolates is completed by the study of the fermentative profile of sugars according to the bibliographic data (Lopez and Mayo, 1994; Mathara et al., 2004; Lee et al., 2006).
The species Lactococcus lactis represents 76% of the total with 84 strains, have growth properties at 10°C and not at 45°C, they grow at 4% NaCl (except Lactococcus lactis. Subsp. cremoris), some subspecies can grow at 0.1% Sherman's milk (Kandler et al., 1968).
Among these isolates, 5 phenotypic profiles were distinguished:
Profile A contains 45 strains: (Lv11, Lv6, Lv50, Lv09, Lv63, Lv3, Lv81, Lv43, Lv32, Lv24, Lv60, Lb22, Lb4, Lb65, Lb89, Lb16, Lb45, Lb19, Lb25, Lb66, Lc6, Lc55, Lc89, Lc46, Lc79, Lc3, Lc32, Lc60, Lc85, Lc26, Lc22, Lc1, Lm95, Lm90, Lm5, Lm36, Lm85, Lm76, Lm54, Lm17, Lm19, Lm23, Lm29, Lm26, Lm49) share the characteristics of growth at 10° and 40°C, grow at 4% NaCl, hydrolyze esculin, do not reduce citrate, do not produce acetoin, hydrolyze arginine and have the ability to grow at 0.1% Sherman's milk, their fermentation profile shows that these strains ferment mainly ribose, mannitol, lactose, starch, maltose, fructose, sucrose do not ferment rhamnose and sorbitol. These isolates belonging to the species Lc. Lactococcus lactis subsp. lactis (Sharpe, 1979; Schleifer et al., 1985; Barlows et al., 1991).
Profile B presents 11 strains (Lv76, Lv29, Lb4, Lb1, Lc72, Lc16, Lc15, Lm39, Lm9, Lm7, Lm82) that do not hydrolyze arginine, do not produce acetoin, do not degrade citrate, do not grow at 4% NaCl, grow at 40°C and hydrolyze esculin, ferment mannitol, D-xylose, do not ferment raffinose nor ribose. These strains may be related to Lactococcus lactis.subsp.Cremoris (Guessas and Adjoudj., 2012; Hadef, 2012; Badis et al., 2014).
Lc. lactis subsp. cremoris is commonly used as a lactic starter and is recognized as the best culture for making "Cheddar" cheese.
Profile C contains 13 isolates (Lv98, Lv78, Lv53, Lb73, Lc29, Lc43, Lc65, Lc33, Lm64, Lm66, Lm57, Lm60, Lm33) that ferment arabinose and mannitol and do not ferment ribose, sorbitol, and raffinose, they are capable of metabolizing citrate. These strains belong to the species Lactococcus lactis subsp. lactis biovar. diacetylactis (Lopez and Mayo, 1994; Badis, 2004; Mathara et al., 2004; Lee et al., 2006).
Among lactococci, only Lc. diacetylactis has the ability to metabolize citrate from milk. The final metabolites from citrate are; diacetyl, acetoin, 2,3-butylene glycol, acetaldehyde, ethanol and lactic acid (Libudzisz and Galewska, 1991), which contributes to the development of flavour in dairy products.
Among these two profiles B and C two subspecies (Lm82, Lm57) isolated from raw camel milk could grow at 6.5% NaCl as reported by Karam (2006).
Camel milk is characterized by a relatively high salinity compared to other milks, which makes it a more or less special environment, this salinity is probably a physiological adaptation of the animal to the desert environment. The bacteria that grow in camel milk are adapted to its salinity and can therefore be exploited in its technological transformation and in the fermentation of salted products such as salted cheese, olives and cucumbers (Zadi-Karam and Karam, 2006).
The D profile corresponds to the Lactococcus lactis subsp hordniae phenotype with 12 strains (Lm72, Lm55, Lc13, Lc9, Lb9, Lb21, Lb65, Lb14, Lv6, Lv62, Lv15, Lv44) that do not grow at 0.1% Sherman's milk, hydrolyze arginine, reduce citrate, produce acetone, do not grow at 4% NaCl, their fermentative profile shows that they do not ferment ribose, D-xylose, mannitol, lactose, maltose, mellibiose, raffinose, except sucrose.
Three (3) of them (Lb49, Lb30, Lm42) belonging to profile E have the phenotype of Lactococcus lactis subsp tructae as they can grow in the presence of 4% NaCl, hydrolyze arginine, ferment ribose, raffinose, mannitol, lactose, mellibiose (Pérez et al., 2011).
The species of Lactococcus lactis and its subspecies are used on a large scale in the dairy industry, they are generally recognized as safe (GRAS) for human consumption. They can be used in the bio-preservation of foods thanks to its ability to produce organic acids and bacteriocins, we take the example of nisin as the most known bacteriocin in the field of food bio-preservation, produced by Lc. lactis. Lactococcus species are also used as probiotic cultures (Kimoto et al., 1999, 2003; Suzuki et al., 2008).
Some isolates (Lm12, Lm11, Lm96, Lm1, Lm44, Lc7, Lc12, Lc59, Lc74, Lc81, Lc25) also do not hydrolyze arginine and produce citrate, are resistant to a concentration of 4% NaCl and a temperature of 40°C, ferment raffinose and do not ferment sorbitol, and can be related to Lactococcus raffinolactis (Badis et al., 2004) (Lopez and Mayo, 1994; Mathara et al., 2004; Lee et al., 2006).
The 11 strains (Lm38, Lm25, Lm20, Lc19, Lc6, Lb80, Lb92, Lb46, Lv54, Lv17, Lv91) do not hydrolyze arginine and produce acetoin, reduce citrate, ferment maltose and not sorbitol, lactose, starch, fructose, arabinose, raffinose These isolates can be related to Lactococcus plantarum (Lopez and Mayo, 1994 Badis et al., 2004; Mathara et al., 2004; Lee et al., 2006).
A strain isolated from raw goat milk (Lc93) has a phenotype of the species Lactococcus piscium, it does not hydrolyze arginine, produces citrate, does not reduce acetoin, does not resist salt concentration higher than 2%. (Williams et al; 1990)
Lastly, arriving the two species Lactococcus hircilactis (Lc47 and Lc95) isolated from raw goat milk (meucci et al., 2015) and Lactococcus laudensis (Lv16) isolated from raw cow milk (Meucci et al., 2015).
Both strains Lc95 and Lc47 resist to grow at pH 4 and not at 4% salt concentration, hydrolyze arginine, ferment galactose, mellibiose, raffinose, sucrose, esculin and not ribose and D-xylose.
The Lv16 strain hydrolyzes arginine, grows at 4% NaCl, does not resist pH4, does not ferment raffinose, ribose, mellibiose.
However, the phenotypic characterization, although it is a fundamental and preliminary step, it remains sketchy and imprecise. Also and in order to assign acceptable and reliable profiles to the selected strains, a molecular identification is essential and mandatory (Ennadir et al., 2014) (Fig 1 and 2).
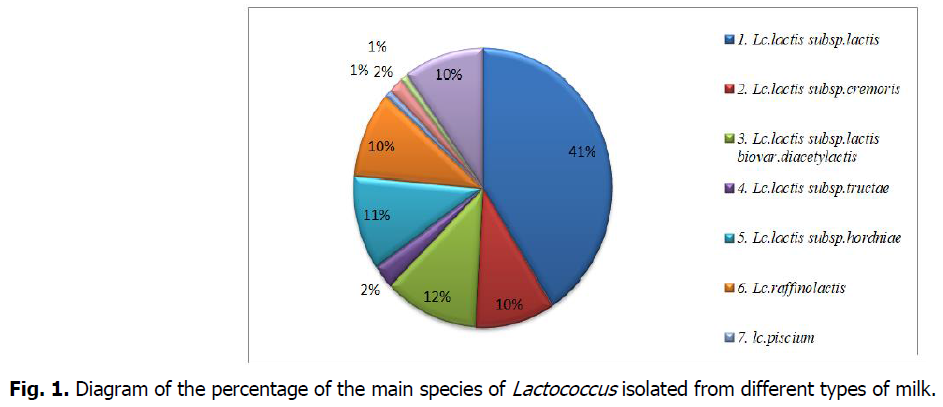
Fig 1: Diagram of the percentage of the main species of Lactococcus isolated from different types of milk.
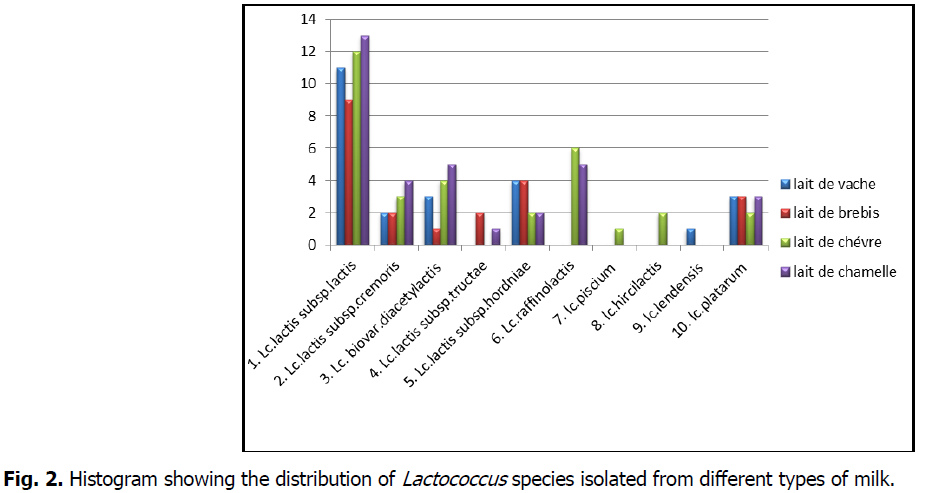
Fig 2: Histogram showing the distribution of Lactococcus species isolated from different types of milk.
Genotypic identification of strains
Based on the biotechnological characteristics of the isolated lactococci, we select the best of them and identify them by the molecular method.
After DNA extraction (CTAB/NaCl wilson method) and amplification of the 16S rDNA gene by PCR, we compared the sequences of all the isolated strains.
Then a purification of the 16S rDNA PCR amplicons is performed as previously mentioned, after arriving at the analysis of these DNA sequences using a Big Dye Terminator V3.1 cycle sequencing kit (Applied Biosystems) and an Applied Biosystems 3130XL capillary DNA sequencer.
The sequences obtained after correction and computer processing with several software using the GenBank DNA databases (http://www.ncbi.nih.gov) (Altschul et al., 1990), and an alignment is performed using the site (https://mafft.cbrc.jp/alignment/server) (Fig 3).
Fig 3: Amplification of the 16S rRNA gene of the examined strains.
Phylogenetic analysis of the 16S rDNA gene sequences was performed using MEGA (molecular evolutionary genetics analysis) software, version 6 (Tamura et al., 2013). This software was also used to construct interactive phylogenetic trees and estimate evolutionary distances based on the method using the Neighbor-Joining method (Weisburg et al., 1991). With 1000 bootstrap values and the Maximum Composite Likelihood (MCL) method to estimate evolutionary distances between all pairs of sequences (Felsenstein, 2004) (Fig 4).

Fig 4: Phylogenetic tree of 16S rRNA gene of strains isolated from raw milk of sheep, cow, camel and goat using Neighbor-Joining method NJ Saitou and Nei, 1987 with Bacillus subtilis as Outgroup.
The tree groups two clades with bootstrap values of 70% and 92%.
The first is composed of two sub-branches: isolates 1, 2, 3, and the reference strains Lactococcus lactis subsp.lactis. biovar. diacetylactis, Lactococcus lactis subsp. hordniae, Lactococcus lactis subsp. Cremoris form the first sub-branch with a boostrap value of 100%. The second sub-branch is formed by isolates identified as Lactococcus sp.
The second clade, with a boostrap value of 92%, the strains (Lm85, Lv50, Lb19, Lv3) identified as lactococcus lactis.
All bootstrap values are significant because they are above 70%, so the tree is robust.
Conclusion
The microflora of raw milk is essentially composed of bacteria. These bacteria can be Gram positive or Gram negative. The technological microflora is useful for the food industry. Among the important lactic groups are the lactococci, which are of great importance on an industrial scale and are widely used in the food industry as starter strains in the production of cheese and fermented milk.
The aim of our study is to establish a collection of 110 Lactococcus strains extracted from raw milk of (goat, cow, camel and sheep).
First, a phenotypic characterization based on morphological and biochemical criteria was performed followed by a genotypic characterization by PCR and sequencing of the gene coding for 16S rRNA using advanced molecular techniques and large database bioinformatics software.
The phenotypic, biochemical, physiological and genotypic identification of lactococci shows a significant diversity of species in the genus Lactococcus.
References
Williams, A.M., Fryer, J.L., Collins, M.D. (1990). Lactococcus piscium sp. nov. a new Lactococcus species from salmonid fish. FEMS Microbiology Letters, 68:109-113.
Meucci, A., Zago, M., Rossetti, L., Fornasari, M.E., Bonvini, B., Tidona, F., Giraffa, G. (2015). Lactococcushircilactis sp. nov. and Lactococcuslaudensis sp. nov., isolated from milk. International Journal of Systematic and Evolutionary Microbiology, 65:2091-2096.
Badis, A., Laouabdia-Sellamii, N., Guetarni, D., Kihal, M.Et Ouzrout, R. (2006). Phenotypic characterization of lactic acid bacteria isolated from raw goat milk of two local goat populations "Arabia and Kabyle". Science Technology, 23:30-37.
Badis, A., Guetarni, D., Boudjema, B.M., Henni, D.E., Kihal, M. (2004). Identification and technological properties of lactic acid bacteria isolated from raw goat milk of four Algerian races. Food Microbiology, 21:579-588.
Balows, A., Truper, H., Dvorkin, M., Harder, W., Schleifer, K. (1991). Manual of clinical microbiology. In the Procaryotes. Edited by Schleifer, K. New York: Springer-Verlag. Bangladesh. World Journal of Microbiology and Biotechnology. 23:125-133.
Chammas, I.G., Saliba, R., Béal, C. (2006). Characterization of the fermented milk “Laban” with sensory analysis and instrumental measurements. Journal of Food Science, 71:S156-S162.
Dellaras, C. (2007). Practical microbiology for the analytical laboratory or health control. Lavoisier Tec et Doc. Paris, pp:476.
Doumandji, A., Hellal, A., Saidi, N. (2010). Purification of bacteriocinea from Lactobacillus acidophilus. Review on Microbiologu and Indian San et Environment, 4:25-47.
Elliker, P.R., Anderson, A.W., Hannesson, G. (1956). An agar culture medium for lactic acid streptococci and lactobacilli. Journal of Dairy Science, 39:1611-1612.
Giraud, E., Champailler, A., Moulard, S., Raimbault, M. (1998). Development of a miniaturized selective counting strategy of lactic acid bacteria for evaluation of mixed starter in a model cassava fermentation. Journal of Applied Microbiology, 84:444-450.
Bettache, G., Fatma, A., Miloud, H., Mebrouk, K. (2012). Isolation and identification of lactic acid bacteria from Dhan, a traditional butter and their major technological traits. World Applied Sciences Journal, 17:480-488.
Guiraud, J.P., Rosec, J.P. (2004). Pratique des normes en microbiologie alimentaire. Afnor.
Hadef, S. (2012). Evaluation of the technological and Probiotic aptitudes of local lactic bacteria. Magister thesis defended on 18-04-2012. University Kasdi Merbah-Ouargla, Algeria.
Hammes, P. (1992). The genera lactobacillus and carnobacterium. The Prokyatyotes, pp: 1535-1594.
Hariri, A., Ouis, N., Sahnouni, F., Djilali, B. (2009). Implementation of the fermentation of some lactic ferments in media based on carob extracts. Review on Microbiology and Indian San et Environment, International Congress BIOMED 1Marrakech, pp:37-55.
Harrigan, W.F., McCance, M.E. (1976). Laboratory methods in food and dairy microbiology. Academic Press Inc. (London) Ltd.
Herrero, M., Mayo, B., Gonzalez, B., Suarez, J.E. (1996). Evaluation of technologically important traits in lactic acid bacteria isolated from spontaneous fermentations. Journal of Applied Bacteriology, 81:565-570.
Fhoula, I., Najjari, A., Turki, Y., Jaballah, S., Boudabous, A., Ouzari, H. (2013). Diversity and antimicrobial properties of lactic acid bacteria isolated from rhizosphere of olive trees and desert truffles of Tunisia. BioMed Research International.
Ennadir, J., Hassikou, R., Al Askari, G., Arahou, M., Bouazza, F., Amallah, L., Amine, S.A., Khedid, K. (2014), Phenotypic and genotypic characterization of lactic acid bacteria isolated from wheat flour from Morocco. Journal of Material and Environmental Science, 5:1125-1132.
Johnson, M.E., Steele, J.L. (2001). Fermented dairy products. Dans Food Microbiology: Fundamentals and Frontiers, pp:651 -664.
Felsenstein, J. (2004). Inferring Phylogenies. Sinauer Assoc. Inc., Sunderland, MA.
Wilson, K. (1987). Preparation of genomic DNA from bacteria. In Current Protocols in Molecular Biology, pp:2-5.
Kandler, O., Weiss, N. (1986). Genus lactobacillus beijerinck 1901, 212AL. In Bergey’s Manual of Systematic Bacteriology, 2:1209-1234.
Karam, H., Karam, N.E. (2006). Lactic acid bacteria in camel milk from Algeria: identification of salt-resistant Lactococcus strains. Tropicultural, 24:153-156.
Katoh, K., Rozewicki, J., Yamada, K.D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics, 20:1160-1166.
Kempler, G.M., McKay, L.L. (1980). Improved medium for detection of citrate-fermenting Streptococcus lactis subsp. diacetylactis. Applied and Environmental Microbiology, 39:926-927.
Kihal, M., Prevost, H., Lhotte, M.E., Huang, D.Q., Diviès, C. (1996). Instability of plasmid‐encoded citrate permease in Leuconostoc. Letters in Applied Microbiology, 22:219-223.
Kimoto, H., Nomura, M., Kobayashi, M., Mizumachi, K., Okamoto, T. (2003). Survival of lactococci during passage through mouse digestive tract. Canadian Journal of Microbiology, 49:707-711.
Kimoto, H., Kurisaki, J., Tsuji, N.M., Ohmomo, S., Okamoto, T. (1999). Lactococci as probiotic strains: adhesion to human enterocyte‐like Caco‐2 cells and tolerance to low pH and bile. Letters in Applied Microbiology, 29:313-316.
Kuraku, S., Zmasek, C.M., Nishimura, O., Katoh, K. (2013). A leaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Research, 41:W22-W28.
LANE. (1991). Rrna sequencing. In Nucleic Acid Techniques in Bacterial Systematics, pp:115-176.
Larpent, J.P. (1990). Technical memento of microbiology. 2nd Edition, Lavoisier Tecet Doc, Paris, pp:471.
Larpent-Gourgaut, M., Michaux, O., Larpent, J.P., Desmasures, N., Desmazeaud, M., Mangin, I., Masson, F., Montel, M.C., Tailliez, P. (1997). Lactic ferments and related bacteria in: "Microbiologie alimentaire". ed. Larpent, Tec Doc, Lavoisier, Paris.
Lee, J.Y., Kim, C.J., Kunz, B. (2006). Identification of lactic acid bacteria isolated from kimchi and studies on their suitability for application as starter culture in the production of fermented sausages. Meat Science, 72:437-445.
Lehninger, A.L. (1981). Biochemistry: molecular basis of cell structure and function. 2nd Ed, Flammarion Médicine-Science.
Libudzisz, Z., Galewska, E. (1991). Citrate metabolism in Lactococcus lactis subsp. lactis var. diacetylactis strains. Food/Nahrung, 35:611-618.
Lopez, S., Mayo, B. (1997). Identification and characterization of homofermentative mesophilic Lactobacillusstrains isolated from artisan starter‐free cheeses. Letters in Applied Microbiology, 25:233-238.
Mathara, J.M., Schillinger, U., Kutima, P.M., Mbugua, S.K., Holzapfel, W.H. (2004). Isolation, identification and characterisation of the dominant microorganisms of kule naoto: the Maasai traditional fermented milk in Kenya. International Journal of Food Microbiology, 94:269-278.
Molin, G. (2008). Lactobacillus plantarum: the role in foods and in human health. In Handbook of Fermented Functional Foods, CRC Press, pp:353-393.
Saitou, N., Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4:406-425.
O’sullivan, L., Ross, R.P., Hill, C. (2002). Potential of bacteriocin-producing lactic acid bacteria for improvements in food safety and quality. Biochimie, 84:593-604.
Ross, R.P., Morgan, S., Hill, C. (2002). Preservation and fermentation: past, present and future. International Journal of Food Microbiology, 79:3-16.
Randazzo, C.L., Caggia, C., Neviani, E. (2009). Application of molecular approaches to study lactic acid bacteria in artisanal cheeses. Journal of Microbiological Methods, 78:1-9.
Wehrmuller, K., Ryffel, S. (2007). Produits au lait de chèvre et alimentation. ALP Actuel. Eds, State Research Agronomy. Liebefeld-Posieux ALP.
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215:403-410.
Samelis, J., Maurogenakis, F., Metaxopoulos, J. (1994). Characterisation of lactic acid bacteria isolated from naturally fermented Greek dry salami. International Journal of Food Microbiology, 23:179-196.
Sanger, F., Nicklen, S., Coulson, A.R. (1977). DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences, 74:5463-5467.
Schleifer, K.H., Kraus, J., Dvorak, C., Kilpper-Bälz, R., Collins, M.D., Fischer, W. (1985). Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. Systematic and Applied Microbiology, 6:183-195.
Sharp, M.E. (1978). The lactic acid bacteria (induding Leuconostocs). XXème Congré de laiterrie. Paris.
Sherman, J.M. (1973). The streptococci. Bacteriological Review, 1:3-97.
Suzuki, K., Funahashi, W., Koyanagi, M., Yamashita, H. (2004). Lactobacillus paracollinoides sp. nov., isolated from brewery environments. International Journal of Systematic and Evolutionary Microbiology, 54:115-117.
Tamime, A.Y. (2002). Microbiology of starter cultures. ROBINSON, RK Dairy Microbiology Handbook, 3:261.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30:2725-2729.
Tania, P., Jose Luis, B., Alvaro, P., Angel, V., Encarna, V., Ignacio, de Blas., Imanol, Ruiz-Zarzuela. (2011). Lactococcus lactis subsp. tructae subsp. nov. isolated from the intestinal mucus of brown trout (Salmo trutta) and rainbow trout (Oncorhynchus mykiss). International Journal of Systematic and Evolutionary Microbiology, 61:1894-1898.
Thomas, T.D. (1973). Agar medium for differentiation of Streptococcus cremoris from the other bacteria. NZJ Dairy Science Technology, 8:70-71.
Weisburg, W.G., Barns, S.M., Pelletier, D.A., Lane, D.J. (1991). 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology, 173:697-703.
Zamfir, M., Vancanneyt, M., Makras, L., Vaningelgem, F., Lefebvre, K., Pot, B., De Vuyst, L. (2006). Biodiversity of lactic acid bacteria in Romanian dairy products. Systematic and Applied Microbiology, 29:487-495.
Author Info
K. Houbad1*, A.M.A. Bekada1, A. Homrani1 and Y. Djellid2,32Laboratory of Biology of Microbial Systems (LBSM), Higher Normal School of Kuba, B.P. 92, Vieux-Kouba, 16308, Alger, Algeria
3Department of Biology, Faculty of Nature and Life Sciences and Earth Sciences, University of Ghardaia, BP 455, 47000, Ghardaia, Algeria
Citation: Houbad, K., Bekada, A.M.A., Homrani, A., Djellid, Y. (2022). Phenotypic and genotypic characterization of lactococci isolated from different kinds of raw milk (goat, cow, sheep and camel). Ukrainian Journal of Ecology. 12:45-60.
Received: 25-Oct-2022, Manuscript No. UJE-22-78203; , Pre QC No. P-78203; Editor assigned: 27-Oct-2022, Pre QC No. P-78203; Reviewed: 07-Nov-2022, QC No. Q-78203; Revised: 12-Nov-2022, Manuscript No. R-78203; Published: 17-Nov-2022, DOI: 10.15421/2022_407
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
