Research Article - (2023) Volume 13, Issue 1
Three peronosporaceae species of the genus Phytopythium menace Nuphar lutea (Sibth and Sm.) colonies occurring in South-mediterranean freshwater wetlands (a rare species in Algeria and Africa)
L. Kachour1,2,3*, H. Alayat2,3 and D. Gacemi-Kirane1Abstract
Oubeira Lake, Northeastern Algeria, is about to undergo critical changes which threaten local aquaplants populations, at the image of Nuphar lutea. Hereby, we took the initiative to identify aquatic microorganisms associated to Nuphar lutea rhizosphere, in order to identify the causes of root-rot disease, observed when sampling plant materials during the early blooming season of the current year. In silico cultures isolation, microscopic characterization and Molecular phylogeny based on rDNA ITS and cox2 sequences of rhizospheric Oomycetes, have been highly significant to distinguish three species from the genus Phytopythium, suspected to be aggressive phytopathogens against Nuphar lutea population in the Oubeira. This waterborne Protist is morphologically Fungi-like, rather Algae-like in its genotype apect, sharing with the sister genus Phytophthora, the role of decomposing biomachines on submerged organic materials within stagnant freshwater ecosystems. Empirical results suspect this aquatic microorganism to develop a pathogenic devastating behavior against the studied helophytes’ fine roots and become so on, the main causal agent of yellow lily extinction from the South Mediterranean area. We remind that Nuphar lutea is an excellent bio-geo-indicator, only recorded in Oubeira, as a rare species in Africa.
Keywords
Oubeira lake, Freshwater, Mediterranean autochtonous populations, Nuphar lutea L., Phytopyhthium, Plant pathogen, Molecular identification.
Introduction
South Mediterranean climate covers a limited number of wetlands and forests, typifying subtropical to tropical ecosystems; they accomplish an agro-ecological key-role facing aridity that continuously extends from the continental Sahara. In Algeria, the National Park of El Kala comprises wetlands which host endemic rare animal and plant species.
In our current study, we are focusing on water lilies, as submerged herbaceous plants to be a true witness of great climate changes in the Mediterranean area. Natives to the northern hemisphere, Water Lilies tend to perish from this region, as the only and unique site of their occurrence all over the African continent (Robinson, 2017), (Ferrez et al., 2014), (Birks, 2013). They are dominantly represented by white water lilies, systematically designated by Nymphaea alba, which recently disappeared from many points within the National Park of El Kala; observational records where noticed, but no studies are yet realized to bring out the real causes of their decay.
In a second range, yellow Nuphar lutea has been at the origin of classifying El Kala among Ramsar sites, since 1980s to date, for being its only and unique propitious ecosystem in Africa, as shown on Fig. 1.
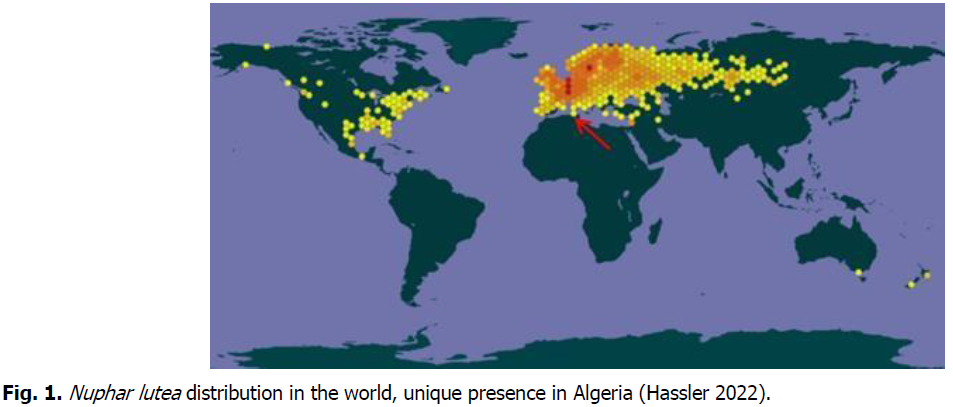
Fig 1: Nuphar lutea distribution in the world, unique presence in Algeria (Hassler 2022).
The species is located in a very limited area that extends to around 500 m2, colonizing the waterway of Oubeira Lake basin and one of its permanent feeding Wadies, called El Kebir, through the so called Daï-Graâ Canal, located at 36°.81842N 8°.44179 E, GPS coordinates (Boumezbeur, 2001), (Betscoun-Muselli, 1989), (Hassler, 2022). Growing in consistent belts, these plants are excellent nests for an important community of fish, amphibians and invertebrates, and perfect habitats for planktonic populations. Some botanists describe Nuphar lutea as one of the eight species of the genus Nymphea, with many synonyms (Betscoun-Muselli, 1989), as listed below:
Basionyme: Nymphaea lutea L. Nenuphar luteum (L.) Link Nymphona lutea (L.) Bubani Nymphozanthus luteus (L.) Fernald.
This Macrofossil is used in ecology as a bio-geo-indicator from the Quaternary age.
It is important to evoke that the species has already disappeared from the Bay of Cannes at the extreme Southeastern Franco-Italian borders, a very similar area, geographically opposite to our site of study Northern the Mediterranean basin (Julve, Ph., 2021). We mention that in Algeria, those flowers still occur in wild contexts and have never been used in artificial ponds, neither in ornamentation nor for biotechnological targets, contrarily to European lilies, which are used for their astringent properties attributed to the tannins present in the rhizome, and the alkaloids of the flowers and rhizomes from both white and yellow waterlilies which have sedative and antispasmodic activity (Betscoun-Muselli 1989).
The problematic loss of such a patrimonial genetic resource can drastically affect the ecosystem. This current study will be the first record of aquatic phytopathogen fungi, which seem to be the causal agents of yellow lily rhizospheric disease.
Materials and Methods
Plant materials sampling
Nuphar lutea plants are preferably growing and colonizing the surface of stagnant shallow freshwater, so that flower, stalks and submerged leaves float at the surface, while the root part emerges on a growing structure of rhizomes strongly incrusted into the sediment floor (Sibth and Sm in Betscoun-Muselli 1989). The rhizosphere is built on branching, spongy, tuberous rhizomes 20-150 mm in diam., toughly attached to the sedimentary mud, so that sampling units were hardly picked at lower depth from five different water input points, along the Daï-Graâ Canal. They were taken from sediments and fine roots materials rotten by aquatic microflora biodegradation, as pictured in Fig. 2.

Fig 2: The Rotten fine roots of Nuphar lutea rhizome, sampled in Oubeira Lake.
Biological materials were kept in clean zipper-bags and immediately transferred to the laboratory in no more than one hour time lapse, at around 7°C could.
Microbial culture and isolation
Microbiological labor consisted in separating fine rotten roots from the rhizome, so that disinfected pieces of about 20 mm length are plated on modified water agar medium, using 9 cm diameter Petri dishes. Water agar was prepared to be selective for Oomycetes by adding to its composition, the following antibiotics: Ampicillin, Nystatin, Pimaricyn, Rifampicyn and Pentachloronitrobenzene (Eckert and Tsao, 1962) (Samson et al., 2004). Resulting mixed colonies with fungiform hyaline soft mycelium, typical to Oomycetes, were isolated after three days to one week incubation time, at room temperature (Erwin and Ribeiro, 1996). 0.5 mm2 Fragments of the juvenile mycelium were isolated on the same modified water agar, as described above. The isolates were cultured at room temperature and incubation took eight to ten days to be ready for microscopic visualization.
Microscopy
Microscopic morphology was obtained by using a photonic microscope BX50 from Olympus Company, and a Malassez-cell to establish lengths scale. Hyaline aseptate hyphae with obovoid to pyriform or curved shaped Sporangia, typical to the Peronosporaceae Oomycetes family, allowed us to consider the following molecular study of the selected isolates.
DNA extraction, amplification and purification
Targeted Oomycetes DNA loci were extracted using GenEluteTM plant Genomic DNA Miniprep Kit (Sigma-Aldrich) according to Manufacturer's specifications. Internal Transcribed Spacer (ITS region) was amplified using the universal primers ITS-LR0 (GCT TAA GTT CAG CGG GT) (Moncalvo et al., 1995), while cytochrome c oxidase subunit II (cox2) was amplified using the primer cox2-F (GGC AAA TGG GTT TTC AAG ATC C) (Hudspeth et al., 2009); we added 1 µg DNA to the reactional medium furnished by Thrmoscientific company; the latter comprises the Master Mix 10 × Dream Taq Green buffer (containing 20 mM MgCl2), 5µL dNTPs (0.2 mM for each nucleotide), primers (0.4 μM of each primer), 1.25 U Taq polymerase; the protocol was realized following the manufacturers’ notice.
Cycling protocol was accomplished by using the following program: step (1) initial denaturing for 3 min at 95°C; step (2) denaturing for 30 sec at 95°C; step (3) annealing for 30 sec at 55°C; step (4) extension for 1 min at 72 °C; step (5) final extension for 5 min at 72°C. The steps 2-4 were repeated 35 times (White et al., 1990). The PCR products were quantified by Electrophoresis on Agarose gel, then finally cleaned-up using Thermo-Fisher-Scientific kit, by adding to 5 µl of it, a mix of 0.5 µl Exonuclease 1 and 1 µl Fast-up alkaline phosphate according to manufaturer's specifications (Ginetti, et al., 2014), then sent for sequencing.
Bioinformatic sequence analyses
Amplicons sequences were checked in Chromas Lite v. 211, then assembled, analyzed and edited in Geneious v.8.1. in a previous step to Geneious analysis, we had to select manually the reference sequences from Genbank database. Multiple alignment was performed using Mafft 7 online aligner, by choosing Q-INS-i strategy setting and 1PAM=2k for scoring matrix. Then, we chose Maximum Parsimony method with default parameters, to obtain the best cladogram from the produced trees (Baldwin, 1992). Bootstrap statistics and branch lengths were computed with Mega 6 (Tamura, et al., 2013).
Results and Discussion
Our results represent the first attempt to identify a Eukaryotic lower aquatic microorganism colonizing the rhizospheric part of Nuphar lutea; it is suspected to be the direct phytopathogen that develops a kind of disease to the smooth tissues of the rhizome, including fine roots.
Molecular identification
Based on molecular results, De Novo assembly allowed us to select 12 specimens of rDNA ITS data of around 700 bp lengths. Over 100 BLAST (NCBI) analyses score permitted to select 12 most similar reference sequences belonging to the genus Phytopythium (Table 1).
| Isolate | Closest reference within Genbank database |
Identity % | Reference source |
|---|---|---|---|
| LK6_LR0 | Phytopythium vexans MT611229.1 | 99.49% | Linaldeddu, B.T. 2020 |
| LK9_LR0 | Phytopythium vexans KT148890.1 | 99.49% | Migliorini, D. 2015 |
| LK10_LR0 | Phytopythium helicoides KT223574.1 | 95.43% | Browne, G.T. 2016 |
| LK11_LR0 | Phytopythium vexans KT148890.1 | 96.33% | Migliorini, D. 2015 |
| LK12_LR0 | Phytopythium vexans OK257570.1 | 84.02% | Panek, M. 2021 |
| LK18_LR0 | Pythium vexans MT611229.1 | 99.50% | Linaldeddu, B. 2020 |
| LK25_LR0 | Phytopythium vexans MF984113.1 | 99.75% | Chen, J.J. 2017 |
| LK27_LR0 | Phytopythium palingenes MN872742.1 | 91.96% | jung 2020 |
| LK34_LR0 | Phytopythium vexans MT611229.1 | 97.06% | Linaldeddu, B.T. 2020 |
| LK35_LR0 | Phytopythium helicoides KT223574.1 | 98.97% | Robideau 2011 |
| LK36_LR0 | Phytopythium vexans MT611229.1 | 99.49% | Linaldeddu, B. 2020 |
| LK46_LR0 | Phytopythium vexans MT611229.1 | 99.37% | Linaldeddu, B. 2020 |
Table 1. Best identity scores with Genebank references acquired by BLAST (ncbi) analysis according to rDNA ITS sequences.
A very high similarity reaching 99.70% with reference sequences from Genebank, allows to highlight three species of the genus Phytopythium, Php. Vexans, Php. helicoides and Php. palingenes. The latter is a newly described species that was isolated from Viet Nam forests by Jung, et al.,2020. Thus, similarity with our isolated species does not exceed 91.96%; better result will be given by Cox2 locus comparison. Sequences of around 530 bp lengths with over 95% similarity to 12 references belonging to the genus Phytopythium, are represented on Table 2.
| Isolate | Closest reference Within Genbank database |
Identity% | Reference source |
|---|---|---|---|
| LK6_cox2 | Phytopythium vexans MH492367.1 | 92.44% | Park, M.J., Back, C. 2019 |
| LK9_cox2 | Phytopythium vexans GU133560.1 | 96.57% | Spies, C.F. 2016 |
| LK10_cox2 | Phytopythium helicoides KT595689.1 | 97.33% | Chen, X.R. 2015 |
| LK11_cox2 | Phytopythium vexans GU133560.1 | 97.30% | Spies, C.F. 2016 |
| LK12_cox2 | Phytopythium vexans GU133560.1 | 97.65% | Spies, C.F. 2016 |
| LK18_cox2 | Pythium vexans GU133560.1 | 98.08% | Spies, C.F. 2016 |
| LK25_cox2 | Phytopythium vexans GU133518.1 | 95.85% | Spies, C.F. 2011 |
| LK27_cox2 | Phytopythium dogmae MF359561.1 | 97.04% | Bennett, R.M. 2017 |
| LK34_cox2 | Phytopythium vexans GU133560.1 | 97.30% | Spies, C.F. 2011 |
| LK35_cox2 | Phytopythium helicoidesKT595689.1 | 96.03% | Chen, X.R. 2015 |
| LK36_cox2 | Phytopythium vexans GU133560.1 | 96.51% | Spies, C.F. 2016 |
| LK46_cox2 | Phytopythium vexans GU133560.1 | 100% | Spies, C.F. 2016 |
Table 2. Best identity scores with Genebank references acquired by BLAST (ncbi) analysis according to cox2 sequences.
Php. Vexans, Php. helicoides and Php. dogmae are the three species of this genus which identify our isolates. We have to justify that based on cox2 results, Php. dogmae presents 97.04% similarity is to our isolate, while ITS sequences comparison to Php. palingenes (Jung, et al.,2020) revealed less similarity to the same isolate. Therefore, we argue our isolate to be rather php. dogmae (Bennett, et al.,2017). Both species Php. palingenes and php. dogmae, were first found in Asian tropics and seem favorably colonizing Mangrove-like wetlands (Jung, et al., 2020), (Bennett, et al., 2017) (Nam, B., and Choi, Y.J., 2019). It is herein identified in the Mediterranean basin as a first record of Php. dogmae in Oubeira Lake, showing their closeness to weather conditions which promote its occurrence in El Kala National park Northeastern Algeria, supposed to be Mediterranean by excellence.
This genus is one of The Peronosporaceae oomycetes, ecologically represented to be phytopathogens in agrarian ecosystems; many of them are known to attack trees roots in forestal ones (Ginetti, et al., 2012). The inventory of native species predominantly occurring in Oubeira inland freshwaters, has revealed a large distribution of the genus Phytopythium species almost identified, as necrotrophic microbs, decomposing dead plant materials and also occurring in sediments and wet soils (Kachour, et al., 2016).
Phylogenetic reconstruction
In light of the software analyses of our resuls, maximum parsimony phylogenetic reconstruction present a cladogram organized into 3 clusters of DNA sequences for both ITS and Cox2 loci, respectively represented on Fig. 3 and Fig. 4.
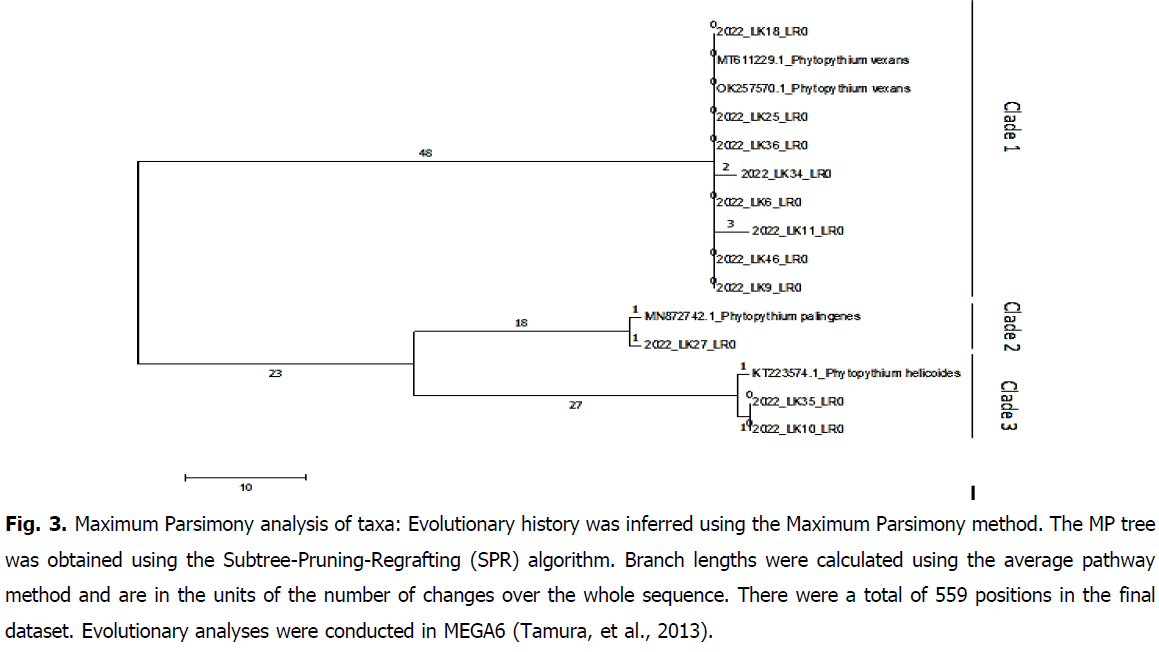
Fig 3: Maximum Parsimony analysis of taxa: Evolutionary history was inferred using the Maximum Parsimony method. The MP tree was obtained using the Subtree-Pruning-Regrafting (SPR) algorithm. Branch lengths were calculated using the average pathway method and are in the units of the number of changes over the whole sequence. There were a total of 559 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Tamura, et al., 2013).

Fig 4: Maximum Parsimony analysis of taxa, Evolutionary history was inferred using the Maximum Parsimony method. The MP tree was obtained using the Subtree-Pruning-Regrafting (SPR) algorithm. Branch lengths were calculated using the average pathway method. There were a total of 468 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Tamura, et al., 2013).
Phylogenetically speaking, Phytopythium has been actualised to count more than 20 sprotrophic or facultative biotrophic genera (Robideau, et al., 2011; Tkaczyk, 2020). Phylogenetic bifurcation into Php. vexans on one hand, and Php. helicoides on the other hand, is given by both ITS and cox2 sequences analyses. The new species Php. dogmae is established to share genetic evolution scenario with the sister species Php. helicoides; microscopic study will be confirmative to support the hypothesis.
Colonies aspect and microscopic morphology
Oomycetes are fungal-like worldwide spread, with the particularity to grow on extremely poor media, such as water agar. Typical isolates with whitish hyaline mycelia growing on modified water agar, as well as morpho-microscopic details, are represented in Fig. 5, Fig. 6 and Fig. 7, respectively corresponding to Php vexans, Php. helicoides and Php. dogmae.
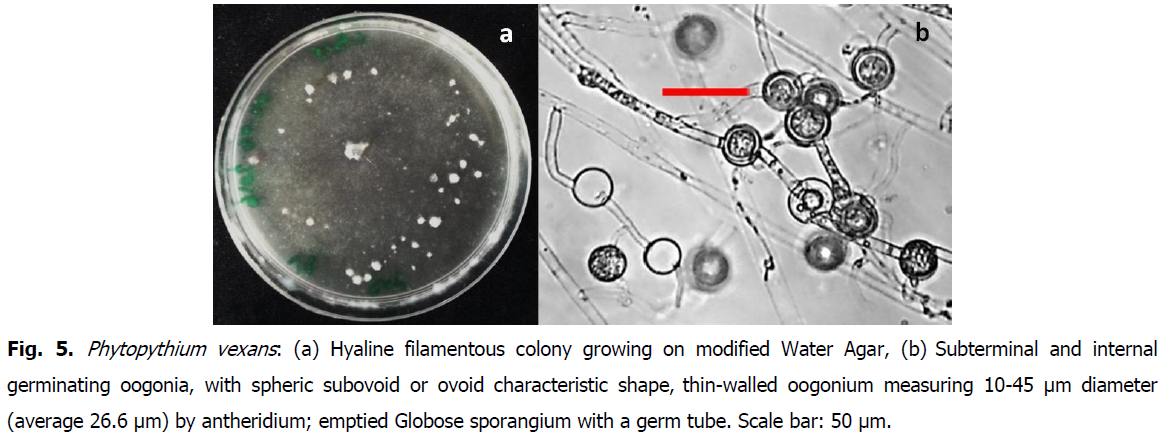
Fig 5: Phytopythium vexans: (a) Hyaline filamentous colony growing on modified Water Agar, (b) Subterminal and internal germinating oogonia, with spheric subovoid or ovoid characteristic shape, thin-walled oogonium measuring 10-45 μm diameter (average 26.6 μm) by antheridium; emptied Globose sporangium with a germ tube. Scale bar: 50 μm.
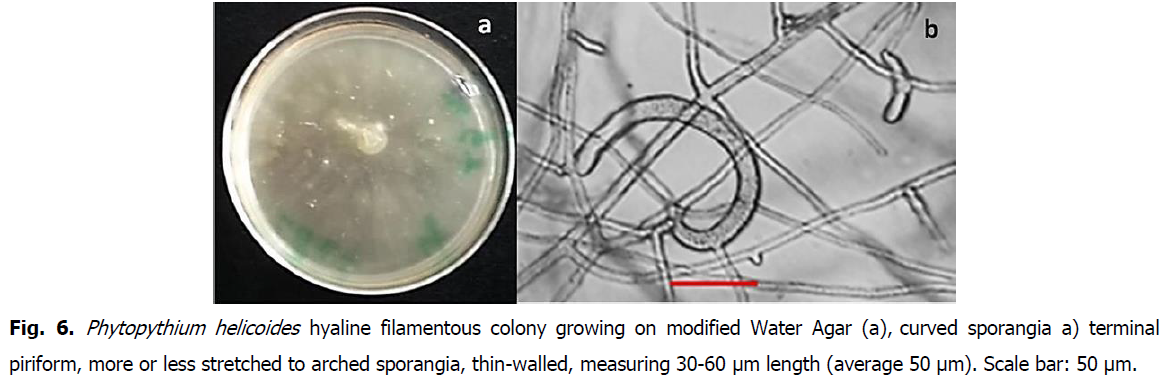
Fig 6: Phytopythium helicoides hyaline filamentous colony growing on modified Water Agar (a), curved sporangia a) terminal piriform, more or less stretched to arched sporangia, thin-walled, measuring 30-60 μm length (average 50 μm). Scale bar: 50 μm.
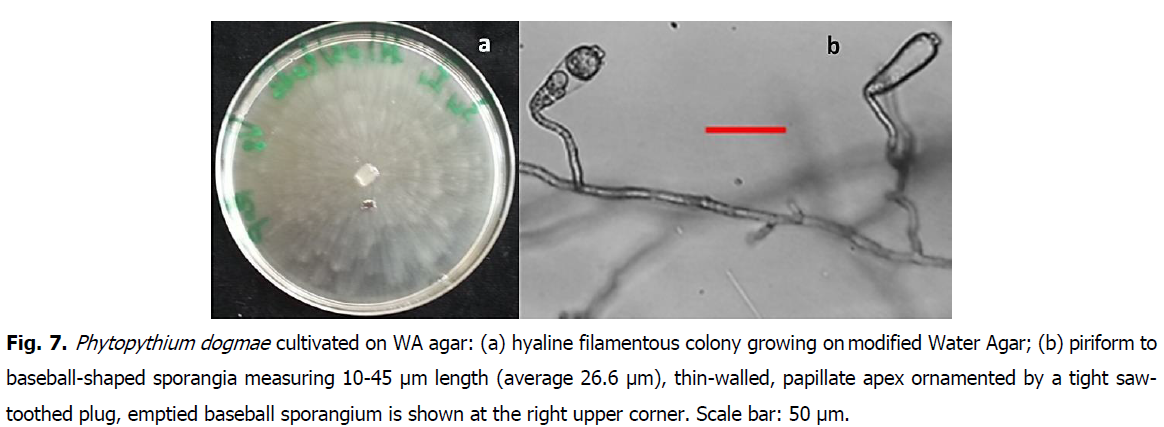
Fig 7: Phytopythium dogmae cultivated on WA agar: (a) hyaline filamentous colony growing on modified Water Agar; (b) piriform to baseball-shaped sporangia measuring 10-45 μm length (average 26.6 μm), thin-walled, papillate apex ornamented by a tight saw-toothed plug, emptied baseball sporangium is shown at the right upper corner. Scale bar: 50 μm.
Frequency
Among the 12 Phytopythium isolates, we could identify 9 specimens which dominantly much to Pph. vexans, and only 2 isolates corresponding to Pph. helicoides; one species was identified as Pph. dogmae, genotypically and phenotypically very similar to the pph. helicoides (Fig. 8). Phytopathological tests were not realized during the current study.
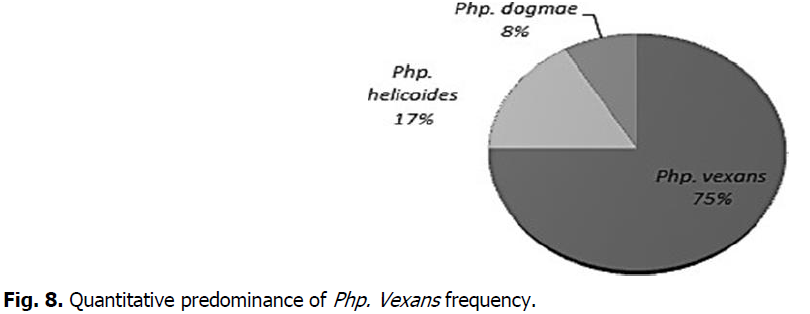
Fig 8: Quantitative predominance of Php. Vexans frequency.
Resistance to the plant immunity
Nuphar luteum biochemical features are diverse, and many chemical compounds showing promise for antifungal, and antibacterial actions, including “alkaloids, terpenoids, saponins, flavonoids, tannins and phenolic compounds, proteins, amino acids, carbohydrates, starch and vitamin C, were listed in many previous reports (Robinson, 2017). Biotechnologically speaking, yellow lilies are widely used in phyto-opharmacology and toxicology as reported in Betscoun-Muselli Isabelle, 1989:
• C15 quinolizidine and piperidine sesquiterpene alkaloids
• Thiaspirane alkaloids in C30
• A glucoside: Nymphaline
These molecules are reported to gather antagonistic properties against pathogens, antibacterial antifungal and antiparasitic virtues. Alcaloïdes are efficient antibacterial antifungal molecules positively tested on phytopathogen bacteria, as Corynebacterium michiganense and many fungi like Histoplasma, Blastomyces, and Tricophyton. Eventhough its biochemical virtuous properties, to elaborate aroma produced by the flowers which is similar to stale alcohol, we could clearly diagnose root rot symptoms on rhizomes and hypothetically suspect oomycetes for being able to escape the biopesticide properties of the elaborated molecules.
Conclusion
The Algerian climate tends to semi-arid, even arid stages, which increasingly causes the scarcity of natural water resources. Along the Tellian-Atlas in the North, numerous inland waterplans still persist and play a vital role against drought. Inside the National Park of El Kala (PNEK) situated at the Tunisian borders, Oubeira Lake, is the largest waterplan, and mainly contributes in annual dynamics maintain of many living populations, such as migrant birds crossing the Mediterranean forth and back. Currently, the lake undertakes a kind of hemorrhagic loss of rare endemic aqua-plants like in our case of study, Nuphar lutea. Three microbial species belonging to the genus Phytopythium are identified to infect fine roots and lead to the plant death; for the first time, Pythium dogme is recorded among the phytopathogens, as a sister pecies with tight similarity to Phytopythium helicoides, with a different behavior and phenetic features.
As the disease is not supposed to be controlled by human interference, the possibility to naturally introduce plant growth promoting rhizobia that may react as an antagonistic agent to the phytopathogen fungi, seems yet, to be one a suggestion to prevent water lily extinction from Oubeira Lake.
References
Bennett, R.M., Nam, B., Dedeles, G.R., Thines, M. (2017). Phytopythium leanoi sp. nov. and Phytopythium dogmae sp. nov., Phytopythium species associated with mangrove leaf litter from the Philippines. Acta Mycologica, 52.
Google Scholar, Crossref, Indexed at
Isabelle, B.M. (1989). Les nénuphars jaune et blanc (Nuphar lutea L. Sibth et Sm, Nymphaea alba L.) (Nymphéacées).
Birks, H.H. (2007). Plant macrofossil introduction. Encyclopedia of Quaternary Science, 3:2266-2288.
Google Scholar, Crossref, Indexed at
Browne, G.T., Schmidt, L.S., Blackburn, N.J. (2016). Pathogenicity of pythium and phytopythium species associated with almond replant disease. Plant Disease.
Boumezbeur, A. (2003). Réserve intégrale du lac oubeira, Wilaya El Tarf. Fiche descriptive sur les zones humides Ramsar. Ministère de l’agriculture et du Développement rural, 7.
Chen, J.J. (2017). Phytopythium aquatile sp. nov. (Pythiaceae, Peronosporales) from southern China based on morphological and molecular. College of Plant Protection, Nanjing.
Eckert, J.W., Tsao, P.H. (1962). A selective antibiotic medium for isolation of Phytophthora and Pythium from plant roots. Phytopathology, 52.
Ferrez, Y., André, M., Gillet, F., Juillerat, P., Philippe, M., Mouly, A., Weidmann, J.M. (2014). Inventaire de la flore vasculaire (Ptéridophytes et Spermaphytes) de Franche-Comté: Indigénats, raretés, menaces, protections. Les Nouvelles Archives de la Flore jurassienne et du nord-est de la France, 11:5-49.
Ginetti, B., Uccello, A., Bracalini, M., Ragazzi, A., Jung, T., Moricca, S. (2012). Root rot and dieback of Pinus pinea caused by phytophthora Humicola in Tuscany, central Italy. Plant Disease, 96:1694-1694.
Ginetti, B., Ragazzi, A., Moricca, S. (2014). First report of Phytophthora taxon walnut in Lombardy, North Italy. Plant Disease, 98:424.
Hudspeth, D.S., Nadler, S.A., Hudspeth, M.E. (2000). A COX2 molecular phylogeny of the Peronosporomycetes. Mycologia, 92:674-684.
Google Scholar, Crossref, Indexed at
https://www.tela-botanica.org/projets/phytosociologie
Jung, T., Scanu, B., Brasier, C.M., Webber, J., Milenković, I., Corcobado, T., Horta Jung, M. (2020). A survey in natural forest ecosystems of Vietnam reveals high diversity of both new and described Phytophthora taxa including P. ramorum. Forests, 11:93.
Google Scholar, Crossref, Indexed at
Kachour, L., Gacemi-Kirane, D., Loucif, L., Alayat, H. (2016). First survey of aquatic microbial Fungi-like Pythiaceae predominantly colonizing the south-Mediterranean freshwater wetlands. Research Journal of Pharmaceutical Biological and Chemical Sciences, 7:3067-3078.
Linaldeddu, B.T., Bregant, C., Ruzzon, B., Montecchio, L. (2020). Coniella granati and Phytophthora palmivora the main pathogens involved in pomegranate dieback and mortality in north-eastern Italy. Italian Journal of Mycology, 49:92-100.
Liu, P.W., Chen, C. (2010). National Chung Hsing University, 250 Kuo Kuang Rd, Taichung 402, Taiwan.
Migliorini, D., Ghelardini, L., Tondini, E., Luchi, N., Santini, A. (2015). The potential of symptomless potted plants for carrying invasive soilborne plant pathogens. Diversity and Distributions, 21:1218-1229.
Nam, B., Choi, Y.J. (2019). Phytopythium and Pythium species (Oomycota) isolated from freshwater environments of Korea. Mycobiology, 47:261-272.
Nechwatal, J., Wielgoss, A., Mendgen, K. (2005). New oomycetes from reed stands of Lake Constance. Phytopathology, Universitaet Konstanz, Universitaetsstr. Konstanz 78457, Germany.
Nei, M., Kumar, S. (2000). Molecular evolution and phylogenetics. Oxford University Press, USA.
Panek, M., Manasova, M., Hanacek, A., Wenzlova, J., Zouhar, M. (2021). Phytophthora, Phytopythium, Pythium and Globisporangium species associated with damages to strawberry plants in plantations in the Czech Republic. Ecology and Diagnostic of Fungal PlantPathogens, Crop Research Institute, Drnovska 507/73, Prague 16106, Czech Republic.
Park, M.J., Back, C.G., Park, J.H. (2019). Occurrence of Phytopythium vexans Causing Stem Rot on Anthurium andraeanum in Korea. The Korean Journal of Mycology, 47:443-446.
Robideau, G.P., De Cock, A.W., Coffey, M.D., Voglmayr, H., Brouwer, H., Bala, K., André Lévesque, C. (2011). DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Molecular Ecology Resources, 11:1002-1011.
Robinson, E. (2017). Nature notes: Water Lilies. Harpswell Heritage Land Trust’s association.
Samson, R.A., Hoekstra, E.S., Frisvad, J.C. (2004). Introduction to food-and airborne fungi. Centraalbureau voor Schimmelcultures (CBS).
Hassler, M. (2021). Synonymic checklists of the vascular plants of the world. Catalogue of Life Checklist (Version 2021-08-06).
Vandepitte, L., Hobern, D., Remsen, D., Schalk, P., DeWalt, R.E., Keping, M., Miller, J., Orrell, T., Aalbu, R. Catalogue of life checklist.
Spies, C.F., Mazzola, M., Botha, W.J., Van Der Rijst, M., Mostert, L., Mcleod, A. (2011). Oogonial biometry and phylogenetic analyses of the Pythium vexans species group from woody agricultural hosts in South Africa reveal distinct groups within this taxon. Fungal Biology, 115:157-168.
Google Scholar, Crossref, Indexed at
Spies, C.F.J., Rintoul, T.L., de Cock, A.W.A.M., Glockling, S.L., Levesque, C.A. (2014). Phylogenetic backbone of the genus Pythium. Agriculture and Agri-Food Canada, Biodiversity, 960 Carling Avenue, Ottawa, ON K1A 0C6, Canada.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30:2725-2729.
Google Scholar, Crossref, Indexed at
Tkaczyk, M. (2020). Origin, differences and meaning in modern plant pathology. Folia Forestalia Polonica, 62:227-232.
Google Scholar, Crossref, Indexed at
Yang, X., Hong, C.X. (2016). Diversity and populations of Phytophthora, Phytopythium and Pythium species recovered from sediments in an agricultural run‐off sedimentation reservoir. Plant Pathology, 65:1118-1125.
Author Info
L. Kachour1,2,3*, H. Alayat2,3 and D. Gacemi-Kirane12Chadli Bendjedid University of El Tarf, PB 73 El Tarf 36000, Algeria
3Laboratory of Agriculture and Ecosystems Functioning (LAEF), Algeria
Citation: Kachour, L., Alayat, H., Gacemi-Kirane, D. (2023). Three peronosporaceae species of the genus Phytopythium menace Nuphar lutea (Sibth and Sm.) colonies occurring in South-mediterranean freshwater wetlands (a rare species in Algeria and Africa). Ukrainian Journal of Ecology. 13:8-16.
Received: 13-Dec-2022, Manuscript No. UJE-22-83262; , Pre QC No. P-83262; Editor assigned: 16-Dec-2022, Pre QC No. P-83262; Reviewed: 29-Dec-2022, QC No. Q-83262; Revised: 04-Jan-2023, Manuscript No. R-83262; Published: 13-Jan-2023, DOI: 10.15421/2023_419
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
